% irbexcept "%", which is meant to represent the prompt on your terminal.
irb(main):001:0>If you input a line, the next prompt will be
irb(main):002:0>
After installing NetCDFruby, cut and paste characters after prompts in the following. Lines that do not start with prompts are the outputs you will see. Italicized characters following "#" are comments attached afterwards for your information.
irb(main):001:0> require "numru/netcdf" # load NetCDFruby
true
irb(main):002:0> file = NumRu::NetCDF.create("test.nc") # create a file
NetCDF:test.nc
irb(main):003:0> dim = file.def_dim("x",3) # create a dimension named "x" and length==3
NetCDFDim:x
irb(main):004:0> v=file.def_var("var","float",[dim]) # create a 1D variable with the 1st dim=="x"
NetCDFVar:test.nc?var
irb(main):005:0> file.close # close the file
nil
irb(main):006:0> print `ncdump test.nc`
netcdf test {
dimensions:
x = 3 ;
variables:
double var(x) ; # "float" for ruby is double in C. Use "sfloat" to define the float of C
data:
var = _, _, _ ; # empty because values have not been set
}
nil
Class method "open":
By default, a file is opened as read-only as with the predefined File class.
Therefore,
file = NumRu::NetCDF.open("hogehoge.nc")
fails unless the file "hogehoge.nc" exists. The class method open takes
two optional variables, mode and share, as its definition is
def NumRu::NetCDF.open(filename,mode="r",share=false)A file is opened as writable if mode is set to "a" or "r+". The file is created if it is not found. Thus the method behaves like NetCDF.create in this case. The second optional argument is whether to use the NC_SHARE mode or not (see the NetCDF documentation for its meaning). The default is false as with the original NetCDF library. Follow the following examples to set these options:
file = NumRu::NetCDF.open("hogehoge.nc","a") # writable, no share
file = NumRu::NetCDF.open("hogehoge.nc","a",true) # writable, share
file = NumRu::NetCDF.open("hogehoge.nc","r",false) # read-only, share
The IO mode follows the predefined File class of Ruby, which is based on
the IO mode of the C language: "r" (read only), "w","w+" (write --
current contents are overwritten (eliminated!)), and "r+","a","a+" (append
-- writable while current contents are preserved). Here, "r+" and "a"
is the same, since NetCDF is not sequential. Also, you can always read
in any mode, so "w" and "w+" are the same. The binary flag "b"
is meaningless (thus "r" and "rb" is the same), since NetCDF is always binary.
As seen above, to give the second argument is mandatory to specify the
third one. Therefore, what seems to be less used is placed later (that
is, we think the share option is the least used).
The contents of file demo1-create.rb:
(*NOTE*: The number at the beginning of each line in what follows is
the line number attached afterwards for reference. It is NOT included in
the source code. To download it, click
here)
1: require "numru/netcdf"
2: include NumRu
3:
4: file = NetCDF.create("test.nc")
5: nx, ny = 10, 5
6: xdim = file.def_dim("x",nx)
7: ydim = file.def_dim("y",ny)
8: tdim = file.def_dim("t",0)
9: require "date"
10: file.put_att("history","created by #{$0} #{Date.today}")
11:
12: x = file.def_var("x","sfloat",[xdim])
13: y = file.def_var("y","sfloat",[ydim])
14: t = file.def_var("t","sfloat",[tdim])
15: v1 = file.def_var("v1","sfloat",[xdim,ydim])
16: v1.put_att("long_name","test 1")
17: v1.put_att("units","1")
18: v2 = file.def_var("v2","sfloat",[xdim,ydim,tdim])
19: v2.put_att("long_name","test 2")
20: v2.put_att("units","1")
21: file.enddef
22:
23: x.put( NArray.float(nx).indgen! )
24: y.put( NArray.float(ny).indgen! )
25:
26: z = NArray.float(nx,ny).indgen!*0.1
27: v1.put(z)
28: v1.put( NArray.float(nx).add!(20), "start"=>[0,2],"end"=>[-1,2])
29: v2.put(z, "start"=>[0,0,0],"end"=>[-1,-1,0])
30: t.put( 0, "index"=>[0])
31: v2.put(-z, "start"=>[0,0,1],"end"=>[-1,-1,1])
32: t.put( 1, "index"=>[1])
33:
34: file.close
35: print `ncdump test.nc`
Here, Line 1 loads the NetCDF library.
v1 = file.def_var("v1","sfloat",["x","y"])
Therefore, the program can be rewritten alternatively as
demo1-create-alt.rb.
To execute the program, type the following line (except for the prompt "%"):
% ruby demo1-create.rbThen, the following will be printed on your command-line terminal because of Line 35.
netcdf test {
dimensions:
x = 10 ;
y = 5 ;
t = UNLIMITED ; // (2 currently)
variables:
float x(x) ;
float y(y) ;
float t(t) ;
float v1(y, x) ;
v1:long_name = "test 1" ;
v1:units = "1" ;
float v2(t, y, x) ;
v2:long_name = "test 2" ;
v2:units = "1" ;
// global attributes:
:history = "created by demo1-create.rb 2001-09-17" ;
data:
x = 0, 1, 2, 3, 4, 5, 6, 7, 8, 9 ;
y = 0, 1, 2, 3, 4 ;
t = 0, 1 ;
v1 =
0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9,
1, 1.1, 1.2, 1.3, 1.4, 1.5, 1.6, 1.7, 1.8, 1.9,
20, 20, 20, 20, 20, 20, 20, 20, 20, 20,
3, 3.1, 3.2, 3.3, 3.4, 3.5, 3.6, 3.7, 3.8, 3.9,
4, 4.1, 4.2, 4.3, 4.4, 4.5, 4.6, 4.7, 4.8, 4.9 ;
v2 =
0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9,
1, 1.1, 1.2, 1.3, 1.4, 1.5, 1.6, 1.7, 1.8, 1.9,
2, 2.1, 2.2, 2.3, 2.4, 2.5, 2.6, 2.7, 2.8, 2.9,
3, 3.1, 3.2, 3.3, 3.4, 3.5, 3.6, 3.7, 3.8, 3.9,
4, 4.1, 4.2, 4.3, 4.4, 4.5, 4.6, 4.7, 4.8, 4.9,
-0, -0.1, -0.2, -0.3, -0.4, -0.5, -0.6, -0.7, -0.8, -0.9,
-1, -1.1, -1.2, -1.3, -1.4, -1.5, -1.6, -1.7, -1.8, -1.9,
-2, -2.1, -2.2, -2.3, -2.4, -2.5, -2.6, -2.7, -2.8, -2.9,
-3, -3.1, -3.2, -3.3, -3.4, -3.5, -3.6, -3.7, -3.8, -3.9,
-4, -4.1, -4.2, -4.3, -4.4, -4.5, -4.6, -4.7, -4.8, -4.9 ;
}
1:require "numru/dcl"
2:require "numru/netcdf"
3:include NumRu
4:include NMath
5:
6:## < create a sample netcdf file >
7:def write_file
8:
9: file = NetCDF.create("test.nc")
10: nx, ny = 21, 21
11: xdim = file.def_dim("x",nx)
12: ydim = file.def_dim("y",ny)
13:
14: x = file.def_var("x","sfloat",["x"])
15: y = file.def_var("y","sfloat",["y"])
16: var = file.def_var("var","float",["x","y"])
17: var.put_att("long_name","test variable")
18: file.enddef
19:
20: vx = NArray.float(nx).indgen! * (2*(Math::PI)*1.5/(nx-1))
21: vy = NArray.float(ny).indgen! * (2*(Math::PI)*1.0/(ny-1))
22: x.put( vx )
23: y.put( vy )
24:
25: sx = sin( vx )
26: sy = sin( vy )
27: z = NArray.float(nx,ny)
28: for j in 0..ny-1
29: z[true,j] = sx * sy[j] + 0.00001
30: end
31:
32: var.put(z)
33:
34: file.close
35: print `ncdump -h test.nc`
36:end
37:
38:## < read the file and plot >
39:def draw_graph
40:
41: file = NetCDF.open("test.nc")
42: vx = file.var("x")
43: vy = file.var("y")
44: vz = file.var("var")
45: x = vx.get
46: y = vy.get
47: z = vz.get
48: file.close
49:
50: #DCL.swlset('ldump',1)
51: DCL.gropn(1)
52: DCL.grfrm
53: DCL.usspnt(x, y)
54: DCL.uspfit
55: DCL.usdaxs
56: DCL.sglset("lsoftf",false)
57: DCL.uegtlb(z, -20) # set the number of levels
58: DCL.uelset("ltone",true)
59: DCL.uetone(z)
60: DCL.udcntz(z)
61: DCL.grcls
62:end
63:
64:###(main)###
65:write_file
66:draw_graph
67:###(main)###
To execute the program, type the following line (except for the prompt "%"):
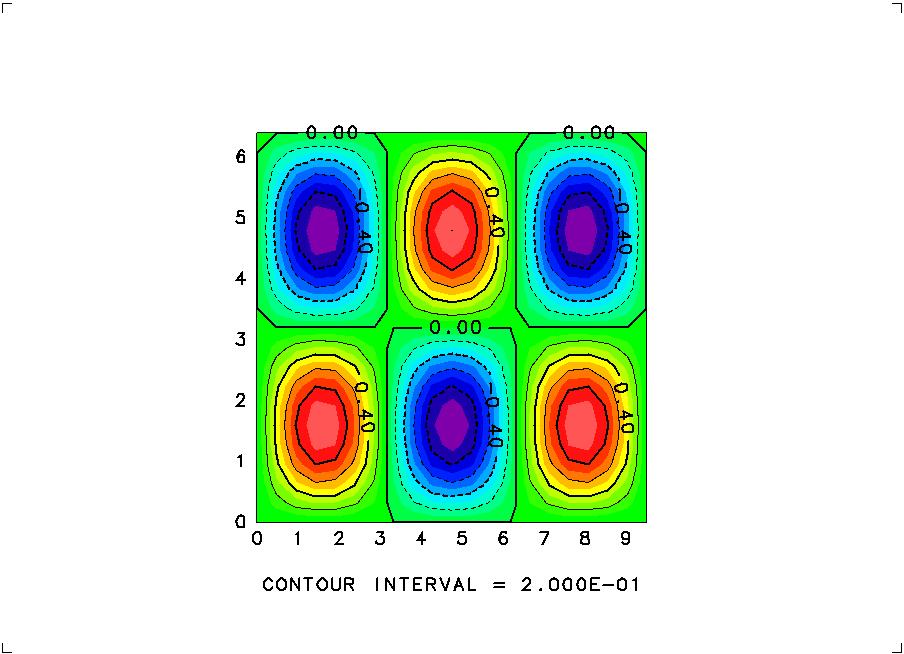
% ruby demo2-graphic.rbThen you will see the next image plotted to appear on your display.

Contents of demo3-ncepclim.rb:
1:# Plot global climatological temperature distribution
2:# from the NCEP reanalysis data.
3:# The data is downloaded if not found and the users wants.
4:
5:######################################
6:### local functions ###
7:def nearest_index(na,val)
8: # returns the element of na nearest to val
9: # na is assumed to be 1d and monotonic
10:
11: if(na[0] > na[-1]) then
12: reversed = true
13: na = na[-1..0]
14: else
15: reversed = false
16: end
17:
18: w = (na.lt(val)).where
19: idx = [ (w.length==0 ? 0 : w.max), na.length-2 ].min
20: if ( na[idx+1]-val < val-na[idx] ) then
21: idx = idx+1
22: end
23:
24: if reversed then
25: na = na[-1..0]
26: idx = na.length-1-idx
27: end
28:
29: idx
30:end
31:#####################################
32:### main part ###
33:require "numru/dcl"
34:require "numru/netcdf"
35:include NumRu
36:
37:filename = "air.mon.ltm.nc"
38:
39:# < download if not found and the users wants >
40:
41:if !(Dir.glob(filename)[0]) then
42: # file not found ==> download by ftp if the user wants
43: host = "ftp.cdc.noaa.gov"
44: path = "/Datasets/ncep.reanalysis.derived/pressure/"+filename
45: print "\n*** question ***\n",
46: " File "+filename+" is not in the current directory.\n",
47: " Would you like to download (ftp) it from "+host+"?\n",
48: " (y, n)> "
49: ans = gets
50: if ans =~ /^y/ then
51: print " What is your email address? (needed for anonymous ftp) > "
52: email = gets.chop!
53: require "net/ftp"
54: print " connecting...\n"
55: ftp = Net::FTP.open(host, "anonymous", email, nil)
56: size = ftp.size(path)
57: print " Size of the file is #{(size/1000)} kb. Would you really like to download?\n (y, n)> "
58: ans = gets
59: if ans =~ /^y/ then
60: print " now downloading...\n"
61: ftp.getbinaryfile(path, filename)
62: else
63: print " exit\n"
64: exit
65: end
66: else
67: print " exit\n"
68: exit
69: end
70:end
71:
72:# < open the file and read axes >
73:
74:file = NetCDF.open(filename)
75:var = file.var("air") # temperature
76:
77:lon = file.var("lon")
78:lat = file.var("lat")
79:level = file.var("level")
80:time = file.var("time") # in hours
81:t = (time.get/720).round + 1 # --> in months
82:axes = [ lon, lat, level, time ]
83:axvals = [ lon.get, lat.get, level.get, t ]
84:axunits = [ lon.att("units").get.gsub("_",""),
85: lat.att("units").get.gsub("_",""),
86: level.att("units").get.gsub("_",""),
87: "months" ]
88:# < graphics >
89:
90:#DCL.sglset('lbuff',false)
91:DCL.swlset('lwait',false)
92:DCL.gropn(1)
93:
94:first = true
95:while true do
96: begin
97:
98: ## / select a 2D slice /
99:
100: if (first) then
101: print <<-EOS
102: ** select a slice **
103: List desired grid numbers of lonigutede, latitude, level, time
104: (set only two of them)
105:
106: Example:
107: , , 850, 1
108: -- horizontal slice at 850hPa and January
109: 135, , , 2
110: -- vertical slice at 135E and Feburary
111: EOS
112: DCL.grfrm
113: else
114: DCL.grfrm
115: print "Input next slice (C-d to quit)\n"
116: end
117: print "> "
118: slice = gets.chop!.split(",")
119: if slice.length!=4 then
120: raise("Slice must be 4 comma-split numerics")
121: end
122: slice.collect!{|e| # "collect!" replaces elements
123: if e =~ /^ *$/ then
124: nil
125: else
126: e.to_f
127: end
128: }
129:
130: iax = []
131: start=[] ; last=[]
132: slice.each_index{|i|
133: if slice[i] == nil then
134: iax.push(i)
135: start.push(0) ; last.push(-1) # from the beginning to the end
136: else
137: idx = nearest_index( axvals[i], slice[i] )
138: start.push( idx ) ; last.push( idx )
139: end
140: }
141:
142: if iax.length != 2 then
143: raise("Specify a 2D slice")
144: else
145: x = axvals[iax[0]]
146: iax[0]==2 ? xr=[x.max, x.min] : xr=[x.min, x.max]
147: xttl = axes[iax[0]].att("long_name").get
148: xunits = axunits[iax[0]]
149: y = axvals[iax[1]]
150: iax[1]==2 ? yr=[y.max, y.min] : yr=[y.min, y.max]
151: yttl = axes[iax[1]].att("long_name").get
152: yunits = axunits[iax[1]]
153: end
154:
155: ## / read the slice and plot /
156:
157: v = var.get("start"=>start, "end"=>last)
158: shp=v.shape; shp.delete(1); v.reshape!(*shp) # delete dims of length==1
159:
160: #Fig.inclpoint(x, y)
161: DCL.grswnd( xr[0], xr[1], yr[0], yr[1] )
162: DCL.grsvpt(0.2,0.9,0.2,0.9)
163: DCL.grstrf
164: DCL.ussttl(xttl," ",yttl," ")
165: DCL.usdaxs
166: DCL.uwsgxa(x)
167: DCL.uwsgya(y)
168: DCL.uelset("ltone",true)
169: DCL.uetone(v)
170: DCL.udcntz(v)
171:
172: first = false
173: rescue
174: print "*Error* ", $!,"\n" # show the error message in ($!)
175: end
176:end
177:
178:DCL.grcls
To execute the program, type the following line (except for the prompt "%"):
% ruby demo3-ncepclim.rbThen you will first see the following questionnaire for downloading. The bold characters are sample input (you must type "y" to download).
*** question *** File air.mon.ltm.nc is not in the current directory. Would you like to download (ftp) it from ftp.cdc.noaa.gov? (y, n)> y What is your email address? (needed for anonymous ftp) >Then it will connect to the server and asks whether you really want to download:
connecting... Size of the file is 8580 kb. Would you really like to download? (y, n)> yType "y" as above to continue. Then you will see this message:
now downloading...It may take a while to download, as the file size is 8.6MB. In the future, it is hoped to minimize downloading, by making possible to transfer only the portion actually needed. When the download is completed, the following message appears on your terminal:
*** MESSAGE (SWDOPN) *** GRPH1 : STARTED / IWS = 1.
** select a slice **
List desired grid numbers of lonigutede, latitude, level, time
(set only two of them)
Example:
, , 850, 1
-- horizontal slice at 850hPa and January
135, , , 2
-- vertical slice at 135E and Feburary
*** WARNING (STSWTR) *** WORKSTATION VIEWPORT WAS MODIFIED.
> 135, , , 2
The messages starting with "***" are from the DCL graphic library, and
you can ignore them. In between are an instruction on how to specify a
slice. Following it, you can type as the bold-faced characters in
above. By "135, , , 2", you have selected the latitude-altitude slice at
longitude 135E and of Feburary. You will then see the following image
to appear on your display.
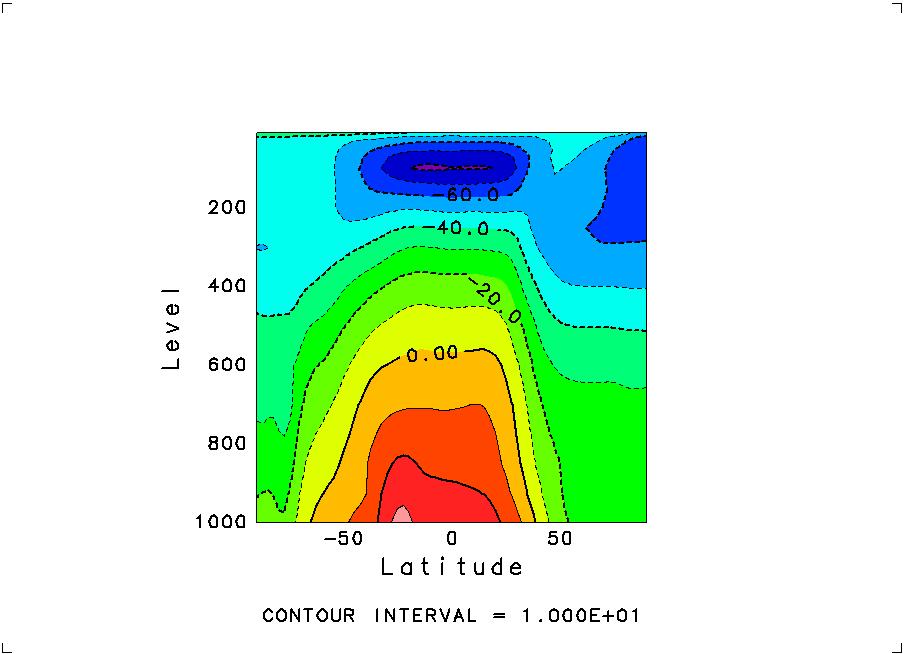
At the same time, you will see the next prompt for another slice as:
*** MESSAGE (UDCNTR) *** INAPPROPRIATE DATA WILL BE MODIFIED INTERNALLY. *** MESSAGE (-CNT.-) *** Z( 7, 7)= -50.0000000 ===> -50.0000610 Input next slice (C-d to quit) *** MESSAGE (SWPCLS) *** GRPH1 : PAGE = 1 COMPLETED. > , , 850, 1Here, " , , 850, 1" was typed in for the longitude-latitude at 850hPa and of January to get the next image:
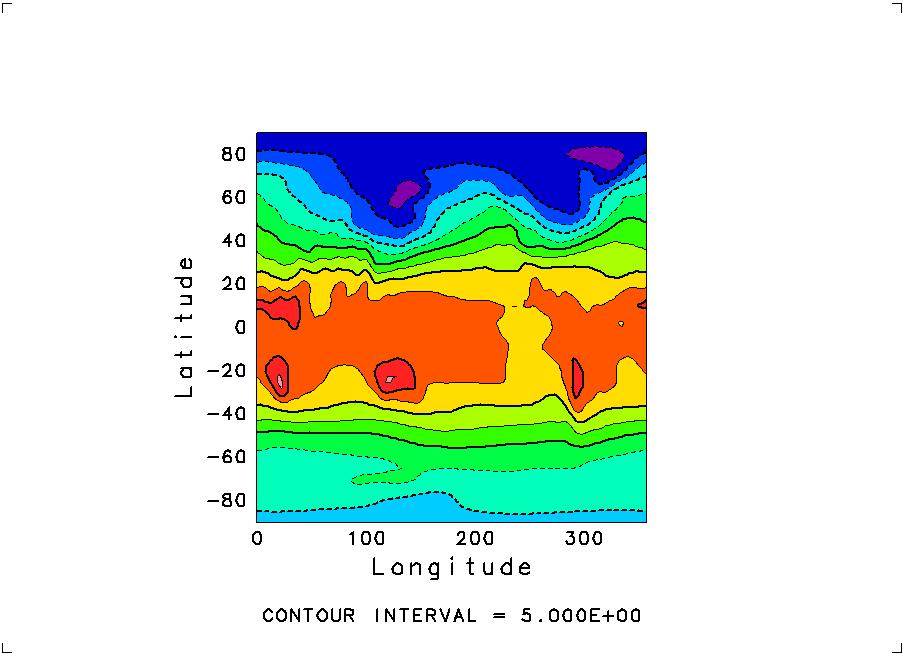
Use Ctrl-d to quit the program.
The following program copies the entire contents of a NetCDF file. Of course, you can simply do it by copying the file (with the cp command, for instance), so it is only to demonstrate how you can handle NetCDF files with this library. The point here is how to use loops.
Contents of demo4-copy.rb:
=begin
=demo4-copy.rb
Make a copy of a NetCDF file
==Usage
% ruby demo4-copy.rb filename_from filename_to
=end
def usage
"\n\nUSAGE:\n% ruby #{$0} filename_from filename_to\n"
end
require "numru/netcdf"
include NumRu
raise usage if ARGV.length != 2
filename_from, filename_to = ARGV
from = NetCDF.open(filename_from)
to = NetCDF.create(filename_to)
from.each_dim{|dim| to.def_dim( dim.name, dim.length_ul0 )}
from.each_att{|att| to.put_att( att.name, att.get )} ## global attributes
from.each_var{|var|
newvar = to.def_var( var.name, var.ntype, var.dim_names )
var.each_att{|att| newvar.put_att( att.name, att.get )}
}
to.enddef
from.each_var{|var| to.var(var.name).put(var.get)}
to.close
Its usage is written above (in the source file). (^_^)
The following program creates a file in the new NetCDF-4 format based on HDF5. Two-dimensional variables in the file are compressed. For comparison, it also creates a file having the same contents but in the traditional NetCDF-3 format.
By default, this library creates file in the NetCDF-3 format. You can change it by calling NetCDF.creation_format=. The expected value is ( NetCDF::NC_NETCDF4 | NetCDF::NC_CLASSIC_MODEL ), or simply NetCDF::NC_NETCDF4. If the former is used, the created file itself is capable of supporting the new data models (such as groups). However, the current ruby-netcdf (ver. 0.7.0) does not support handgling of the new data models.
Contents of demo5-netcdf4.rb:
require "numru/netcdf"
include NumRu
file3 = NetCDF.create("test_nc3.nc")
NetCDF.creation_format = ( NetCDF::NC_NETCDF4 | NetCDF::NC_CLASSIC_MODEL )
file4 = NetCDF.create("test_nc4.nc")
nx, ny = 100, 50
[ file3, file4 ].each do |file|
xdim = file.def_dim("x",nx)
ydim = file.def_dim("y",ny)
x = file.def_var("x","sfloat",[xdim])
y = file.def_var("y","sfloat",[ydim])
v1 = file.def_var("v1","sfloat",[xdim,ydim])
v2 = file.def_var("v2","int",[xdim,ydim])
if /nc4/ =~ file.path
v1.deflate(2) # set the deflation (compression) level 2
v2.deflate(2,true) # set the deflation level 2 with the shuffle filter
puts "v1 deflate params:"
p v1.deflate_params
puts "v2 deflate params:"
p v2.deflate_params
end
file.enddef
x.put( NArray.float(nx).indgen! )
y.put( NArray.float(ny).indgen! )
z = NArray.float(nx,ny).indgen! + 1000000
z[true,ny/2..-1] = 0 # to see the impac
v1.put(z)
v2.put(z)
file.close
end
puts "Created test_nc3.nc test_nc4.nc.", "File size comparison:"
print `ls -l test_nc3.nc test_nc4.nc`
Execute this program as follows:
$ ruby demo5-netcdf4.rbIt then outputs the following:
v1 deflate params: [false, true, 2] v2 deflate params: [true, true, 2] Created test_nc3.nc test_nc4.nc. File size comparison: -rw-r--r-- 1 horinout horinout 40808 Jan 27 20:07 test_nc3.nc -rw-r--r-- 1 horinout horinout 16103 Jan 27 20:07 test_nc4.nc
The last two lines show that the size of the compressed NetCDF-4 file (test_nc4.nc) is much smaller than that of the NetCDF-3 file (test_nc3.nc). Note that the compression rate depends on contents; it is high here because a half of the 2D data is padded with a constant value.
 )
)